Edge Analytics in Healthcare #
Cancer Prediction System
This document provides an introduction to edge analytics in healthcare with SUSE Rancher through the deployment of the Cancer Prediction System. This is an open source, machine learning solution designed to aid medical personnel interpret computer tomography (CT) medical images.
This project was developed as part of the Google Summer of Code (GSOC), a global online mentoring program to introduce new contributors to open source software development, under the guidance of SUSE mentors: Bryan Gartner, Ann Davis, Brian Fromme, and Terry Smith.
Documents published as part of the series SUSE Technical Reference Documentation have been contributed voluntarily by SUSE employees and third parties. They are meant to serve as examples of how particular actions can be performed. They have been compiled with utmost attention to detail. However, this does not guarantee complete accuracy. SUSE cannot verify that actions described in these documents do what is claimed or whether actions described have unintended consequences. SUSE LLC, its affiliates, the authors, and the translators may not be held liable for possible errors or the consequences thereof.
1 Introduction #
1.1 Motivation #
Cancer is the world’s second leading cause of death in the world. It results in development of abnormal cells that divide uncontrollably and can infiltrate and destroy normal body tissue. Survival rates are improved through better screening, treatment, and prevention.
Advanced analytics and machine learning (ML) continue to have a growing influence on modern medicine, accelerating diagnosis, developing treatments, and improving results. Analytics in healthcare is not only taking place in data centers. Increasingly, analytics and ML are deployed at the edge - in clinics, in CT imaging systems and other devices, and even in mobile computing devices, like your doctor’s tablet. This is made possible with modern container technologies, like Kubernetes, which is a portable, extensible, open source platform for managing containerized workloads and services.
In medicine and many other fields, analytics at the edge makes sense. This is where the data, so critical to analytics and ML, is located. And this is where medical professionals interact with patients. This is where they save lives.
This project, resulting in the Cancer Prediction System, is part of the 2022 Google Summer of Code, a global mentoring program focused on introducing new contributors to open source software development. Google Summer of Code contributors are paired with mentors from open source organizations, such as SUSE. They learn from these experienced open source developers, gain exposure to real-world software development techniques, and write code for real-world challenges. This project was developed under mentorship of Bryan Gartner, Ann Davis, and Brian Fromme of SUSE.
This document reviews the approach, implementation, and deployment of this ML-based, Cancer Predicition System, designed to assist medical professionals in early screening.
1.2 Scope #
This guide introduces the basic concepts and steps to install, configure, and use the Cancer Prediction System in a SUSE Rancher Kubernetes environment.
1.3 Audience #
This guide is intended for data scientists, data engineers, and data analysts. It also addresses radiologists, doctors, and hospital administrators who are interested in learning more about the technologies that enable analytics and ML at the edge, particularly for medical use cases. To get the most out of this guide, you should have basic familiarity with the Linux command line and Kubernetes concepts and tooling.
2 Technical overview #
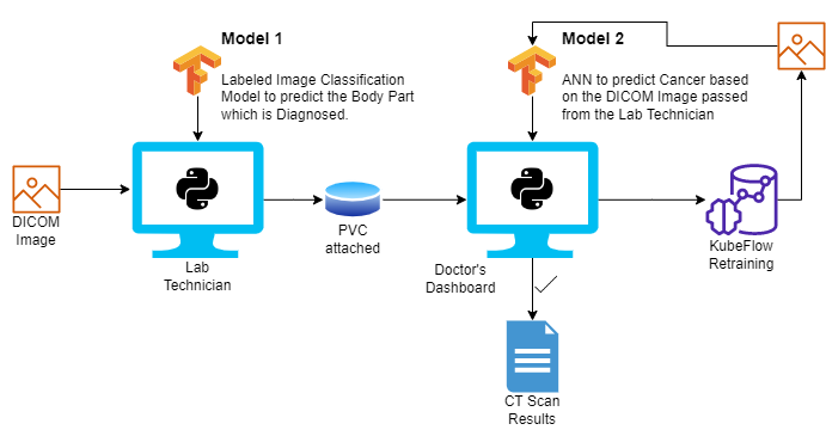
Cancer Predicition System is an ML-based application stack with the goal to assist in cancer risk assessment by predicting the likelihood of developing a cancer prior to occurence of the disease. The image below illustrates the general architecture of the Cancer Prediction System.
The Cancer Prediction System uses CT scan images in the Digital Imaging and Communications in Medicine (DICOM) standard. The system is designed to be deployed in a microservices architecture and is divided into four interfaces:
Lab Technician Dashboard
Doctor Dashboard
ML Pipeline Dashboard
Rancher Dashboard
2.1 Deployment architecture #
The Cancer Prediction System supports a broad spectrum of deployment architectures.
For this guide, the Cancer Prediction System is deployed on a lightweight Kubernetes cluster, featuring K3s 1.21.3+k3s1. The cluster is managed by Rancher 2.6, and persistent storage is provided through Longhorn 1.3.1.
All nodes have the following characteristics:
Processor: 2 vCPUs
Memory: 4 GB RAM
Storage: 10 GB (available)
Operating system: SUSE Linux Enterprise Server 15 SP3
2.2 Components and tools #
The Cancer Prediction System uses a variety of components and tools.
Some highlights include:
- Docker
Docker is a platform that enables developers to build, deploy, run, and manage application containers. This guide uses Docker 20.10.7.
- Flask
Flask is a lightweight Web framework written in Python to easily create Web applications. Flask is used to serve the back-end and ML models as APIs.
- Helm
Helm is a package manager for Kubernetes that helps you define, install, upgrade, and share Kubernetes applications.
- K3s
K3s is a lightweight, CNCF-certified Kubernetes distribution built for IoT and edge computing.
- Keras
Keras is an open source software library that provides a high-level, Python API for designing artificial neural networks. It acts as an interface to the TensorFlow 2 framework. Keras is used to implement the Convolutional Neural Nets (CNNs) in the Cancer Prediction System.
- Kubectl
Kubectl is a command line tool for communicating with a Kubernetes cluster’s control plane via the Kubernetes API.
- Kubeflow
Kubeflow is dedicated to making deployments of ML workflows on Kubernetes simple, portable, and scalable. It is used to define ML pipelines and to orchestrate workflows
- Kustomize
Kustomize is a template-free way to programmatically customize Kubernetes objects and is both a stand-alone tool and built into Kubectl.
- Longhorn
Longhorn is a cloud-native, distributed, persistent block storage system for Kubernetes.
- Manifests
A manifest is a text file (in JSON or YAML format) that specifies the desired state of an object that Kubernetes will maintain. Manifests are used by Kustomize and can be applied with Kubectl.
- SUSE Rancher
Rancher is an enterprise-grade, Kubernetes cluster management platform.
2.3 Process #
Getting started with the Cancer Prediction System is fairly easy. In general, the process is as follows:
Prepare your Kubernetes cluster.
Clone the project repository.
git clone https://github.com/abhi-bhatra/ct_image_scanning.git
Create or update the Kubernetes manifests to define the application environment and deployment.
Log in to your SUSE Rancher environment and select the Kubernetes cluster that will host the application.
Apply Kubernetes manifests to set up the environment (namespaces and storage).
Apply Kubernetes manifests to deploy the application.
Access the application via the Rancher Dashboard.
3 Prepare the Kubernetes cluster #
3.1 Access the Rancher environment #
It is assumed that you already have access to deploy applications to a Kubernetes cluster. If you need to set up your cluster environment, these references may help:
When you are ready, log in to the Rancher UI with your Web browser.
3.2 Install Longhorn #
Before proceeding, review the concepts of persistent volumes, persistent volume claims (PVCs), and storage classes in the Rancher documentation.
Longhorn is a lightweight distributed block storage solution for Kubernetes that is easy to use, self-healing, and highly available. With Longhorn, you can:
partition your block storage into persistent volumes for stateful applications.
replicate block storage across multiple nodes and data centers to increase availability.
schedule recurring snapshots of a volume and recurring backups to external storage.
restore volumes from backup.
create cross-cluster disaster recovery volumes.
Install Longhorn into your Kubernetes cluster using one of the following three methods:
kubectl apply -f https://raw.githubusercontent.com/longhorn/longhorn/v1.3.1/deploy/longhorn.yaml
helm repo add longhorn https://charts.longhorn.io helm repo update helm install longhorn/longhorn -name longhorn -namespace longhorn-system
After successfull installation, you can use the Longhorn Dashboard to manage and monitor the cluster’s Longhorn volumes. Access the Longhorn Dashboard through the Rancher UI.
In the Rancher UI, select the cluster where Longhorn is installed.
In the left navigation menu, click Longhorn.
Click Longhorn in the Overview section to access the Longhorn UI.
3.3 Provision persistent storage #
You can provision persistent storage for your applications with kubectl or by using the Longhorn UI.
See the Longhorn documentation for additional details.
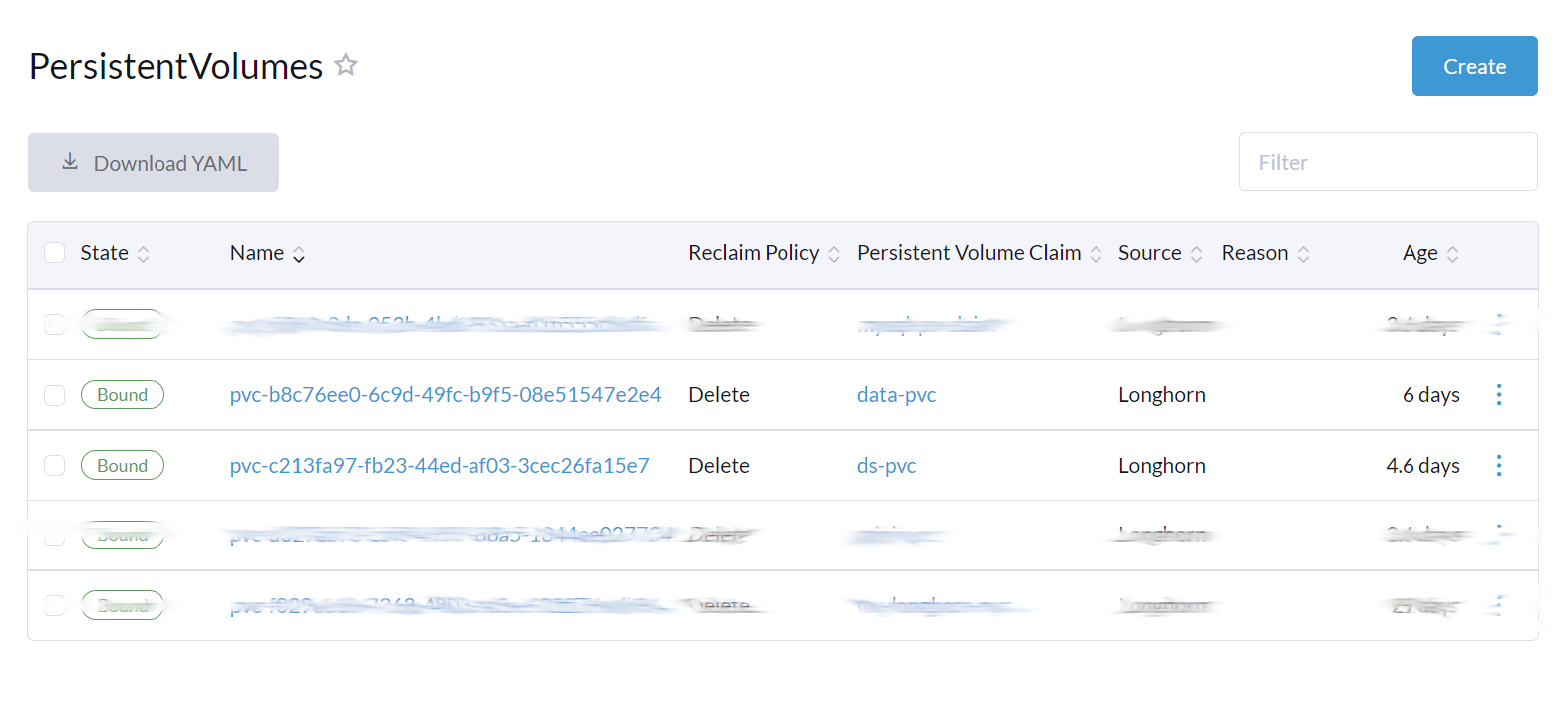
The steps below leverage kubectl to provision PersistentVolumeClaims for two PersistentVolumes.
Create a PersistentVolumeClaim for the applications to share data and information in the cluster.
Create the file 'data-pvc.yaml'.
apiVersion: v1 kind: PersistentVolumeClaim metadata: name: data-pvc namespace: cancerns spec: accessModes: - ReadWriteMany resources: requests: storage: 1Gi storageClassName: longhornImplement this PersistentVolumeClaim.
kubectl apply -f data-pvc.yaml
Create a PersistentVolumeClaim for the data volume.
Create the file 'ds-pvc.yaml'.
apiVersion: v1 kind: PersistentVolumeClaim metadata: name: ds-pvc namespace: cancerns spec: accessModes: - ReadWriteMany resources: requests: storage: 1Gi storageClassName: longhornImplement this PersistentVolumeClaim.
kubectl apply -f ds-pvc.yaml
You can review the persistent volumes in the Rancher UI.
3.4 Install Kubeflow Pipelines #
The Kubeflow project provides a straightforward way to deploy and automate ML workflows on Kubernetes, making it simple, portable, and scalabe. Anywhere you are running Kubernetes, you should be able to run Kubeflow. The Kubeflow documentation provides detailed instructions to install Kubeflow on Kubernetes for production deployments.
Typically, Kubeflow is installed into a large production environment, where it can rely on shared services, like Istio service mesh. For this guide, you can use Kubeflow Pipelines. This stand-alone implementation of Kubeflow does not dependend on other services, providing a simpler installation process.
Kubeflow Pipelines does not support multi-user separation and may not be suitable for production environments.
Set the PIPELINE_VERSION and apply the Kubeflow Pipelines manifest:
export PIPELINE_VERSION=1.8.3 kubectl apply -k "github.com/kubeflow/pipelines/manifests/kustomize/cluster-scoped-resources?ref=$PIPELINE_VERSION" kubectl wait --for condition=established --timeout=60s crd/applications.app.k8s.io kubectl apply -k "github.com/kubeflow/pipelines/manifests/kustomize/env/platform-agnostic-pns?ref=$PIPELINE_VERSION"
NoteIt can take several minutes to deploy Kubeflow Pipelines on your cluster.
Check the status of the deployment by watching the output of:
kubectl get all -n kubeflow
STATUS of all pods is 'Running' once all services have started.
NAME READY STATUS RESTARTS AGE pod/workflow-controller-5667759dd7-fbgrp 1/1 Running 0 2d3h pod/ml-pipeline-scheduledworkflow-7f8bc78db9-qpx4f 1/1 Running 0 2d3h pod/ml-pipeline-viewer-crd-8497d9695c-tqmdg 1/1 Running 0 2d3h pod/ml-pipeline-ui-69bc756bd7-nmzm6 1/1 Running 0 2d3h pod/metadata-envoy-deployment-6df8bdd989-lc77p 1/1 Running 0 2d3h pod/minio-5b65df66c9-qt6lk 1/1 Running 0 2d3h pod/ml-pipeline-persistenceagent-585c4b58d6-mcmtx 1/1 Running 1 2d3h pod/ml-pipeline-7cc4f8fdf7-b2vjp 1/1 Running 2 2d3h pod/cache-server-6cddbbc849-bnd6n 1/1 Running 1 2d3h
Verify the installation by accessing the Kubeflow Pipelines dashboard.
On the command line, create a port forward for the 'ml-pipeline-ui' service.
kubectl port-forward -n kubeflow svc/ml-pipeline-ui 8080:80
Log in to the Rancher UI with your Web browser.
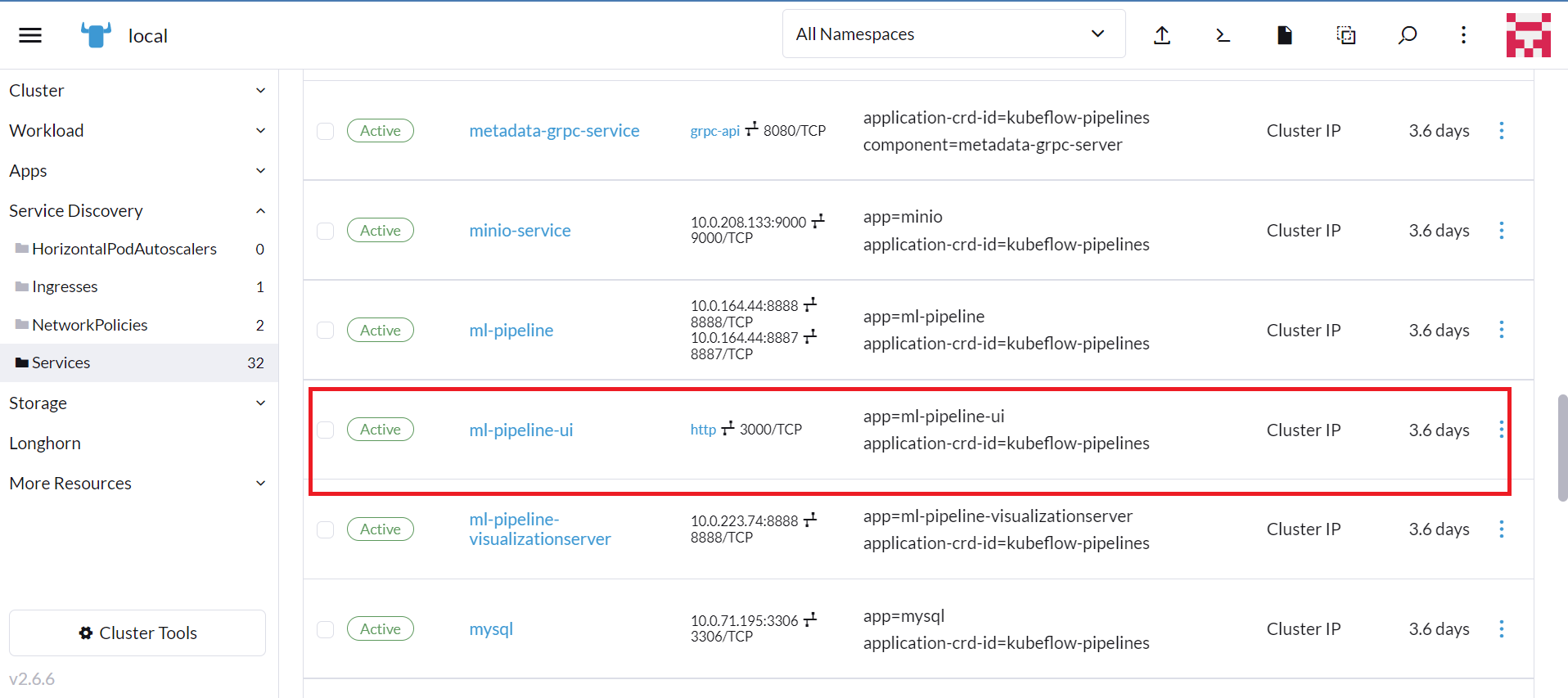
Select your cluster, then click Services.
Click the port indicated for the 'ml-pipeline-ui' service.
Your browser should open to the Kubeflow Pipelines dashboard.
4 Download the data #
The Cancer Imaging Archive (TCIA) is a service that de-identifies and hosts a large archive of medical images of cancer accessible for public download.
Albertina, B., Watson, M., Holback, C., Jarosz, R., Kirk, S., Lee, Y., … Lemmerman, J. (2016). Radiology Data from The Cancer Genome Atlas Lung Adenocarcinoma [TCGA-LUAD] collection. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2016.JGNIHEP5
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. (paper)
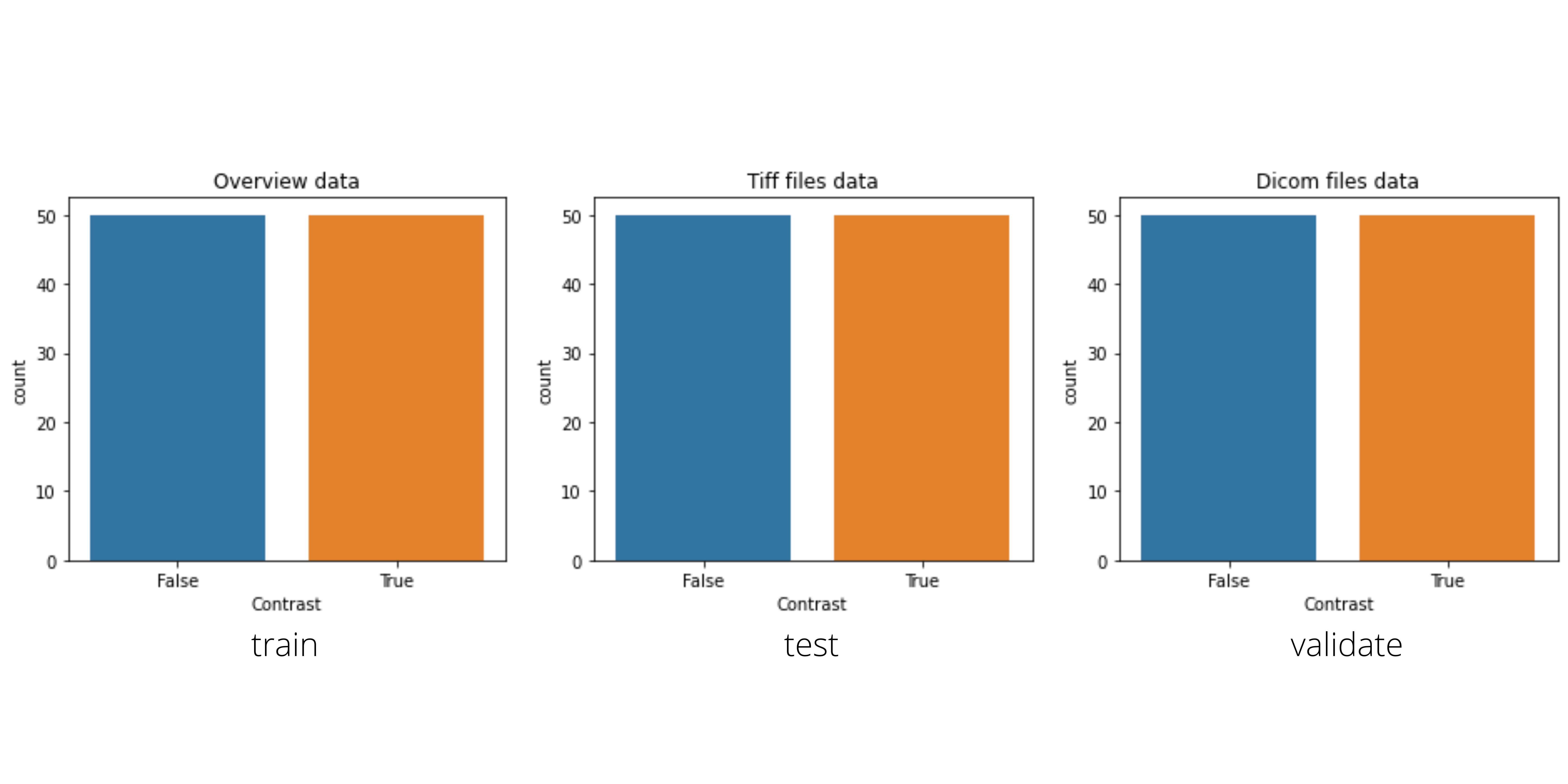
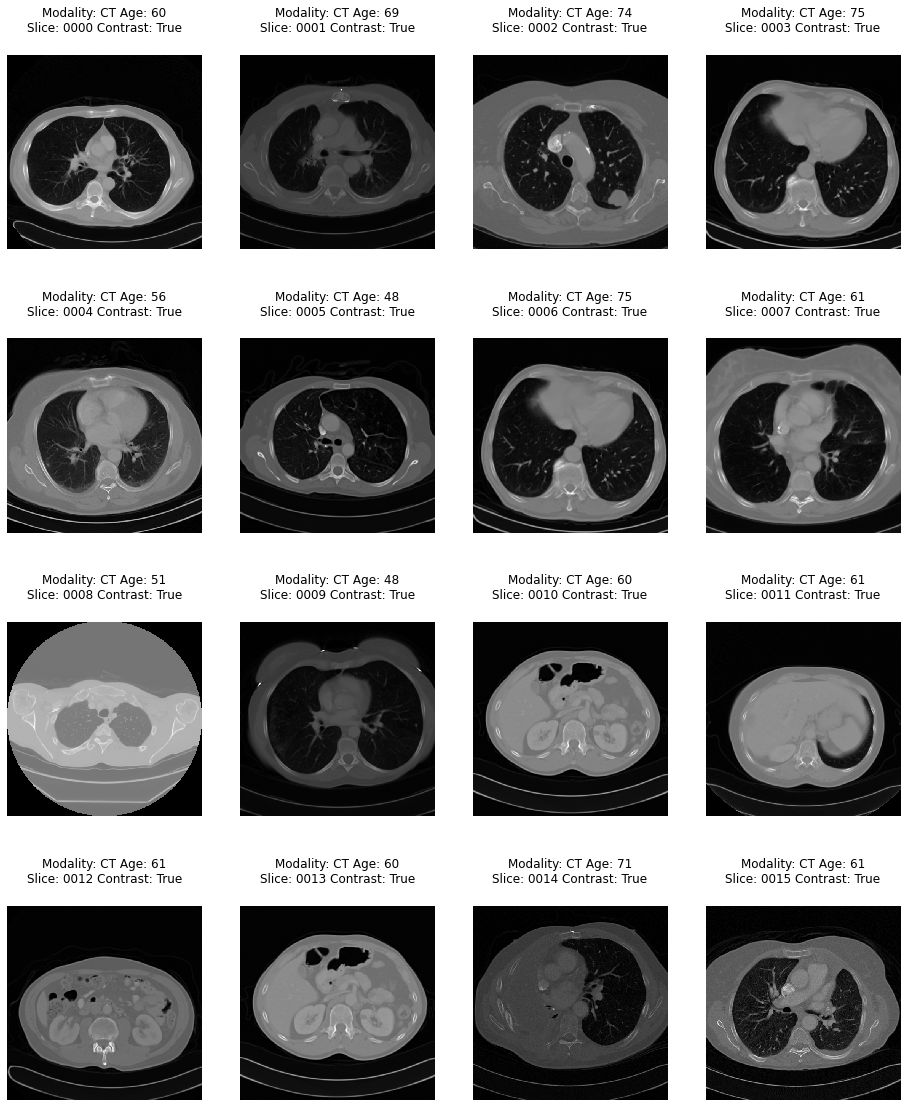
The Cancer Prediction System uses the subset of TCIA images that were featured in a Kaggle competition. This dataset is designed to allow testing of different methods for examining trends in CT image data associated with using contrast and patient age. It consists of 475 series of CT images from 69 participants where valid age, modality, and contrast tags could be found.
Download the dataset from https://www.kaggle.com/datasets/kmader/siim-medical-images as a 262 MB ZIP archive.
NoteYou need a free Kaggle account to download the dataset.
Extract the archive and examine its directory structure and contents.
/dataset -- archive/ -- dicom_dir/ ... ID_0001_AGE_0069_CONTRAST_1_CT.dcm ... -- tiff_images/ ... ID_0000_AGE_0060_CONTRAST_1_CT.tif ... -- full_archive.npz -- overview.csv -- dataset-classification -- Chest-CT/ -- NonChest-CT/ -- dataset-prediction/ -- train/ -- cancer/ -- non-cancer/ -- test/ -- cancer/ -- non-cancer/ -- validation/ -- cancer/ -- non-cancer/archive: Contains the raw dataset downloaded from Kaggle. This is further processed for use in the machine learning model.
dataset-classification: Contains a separate dataset, where all the DICOM images are classified as chest and non-chest. Only chest DICOM images are used for this model, so non-chest images must be filtered out.
dataset-prediciton: Contains all the images separated into train, test, and validation subsets. Each image is labeled as 'cancer' or 'non-cancer'.
Now apply the dataset deployment manifests to download and extract the dataset into the data volume on your cluster.
apiVersion: apps/v1 kind: Deployment metadata: name: datasetvm namespace: cancerns spec: replicas: 1 selector: matchLabels: app: datasetvm template: metadata: labels: app: datasetvm spec: containers: - name: datasetvm image: "ubuntu:latest" imagePullPolicy: Always volumeMounts: - name: dataset mountPath: /dataset env: - name: DATASET value: "https://rancherdataset.blob.core.windows.net/ct-images/dataset.zip" command: ["/bin/sh","-c"] args: ["apt-get update; apt-get install unzip wget -y; wget $DATASET -O /dataset/dataset.zip; unzip /dataset/dataset.zip -d /dataset/dataset; ls -l /dataset/dataset"] volumes: - name: dataset persistentVolumeClaim: claimName: ds-pvc
5 Machine learning model #
The Cancer Prediction System aims to assist a doctor in identifying the probability of cancer in a patient based on a CT image. That is, the Cancer Prediction System must classify a CT image as indicative of cancer or not indicative of cancer. This classification problem can be a challenge for traditional solution methods.
To approach this problem, the Cancer Prediction System uses a predictive model created with a convolutional neural network (CNN). CNNs have been shown to achieve higher accuracy in detecting and segmenting specific objects in images. In this solution, you use a CNN architecture consisting of three convolutional layers and two dense layers with the RMSprop optimizer and binary cross-entropy loss function.
CNNs must be trained before they can be useful. CNN training can involve unsupervised learning or supervised learning. Unsupervised learning is indicated when the categories are unknown or when the training dataset is not already categorized (that is, labeled or tagged). In this case, you have two categories already defined ('cancer' and 'not cancer') and a categorized training dataset. So, the Cancer Prediction System uses supervised learning to train its CNN.
The training process is detailed as follows:
Split the dataset
Split the dataset such that 80% is used for training, 10% is used for validation, and the remaining 10% is used for testing. These images and their attached labels are saved to the
datasetdirectory.Preprocess the data
Convert the images to grayscale and resize to 512x512 pixels, and convert the labels to one hot encoding. Save preprocessed data to the
preprocesseddirectory.Train the model
Feed the training data (images and labels) to the CNN in batches of 32 and do this for 200 epochs. When completed, save the trained model to the
trained-modeldirectory. The validation data can be used to assess the performance of the model and determine whether further training is required.
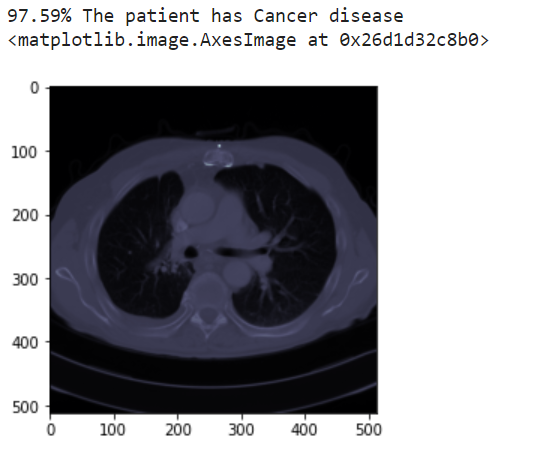
Use the model
Copy the trained model to the
prediction-modeldirectory. Feed a new image (from the test data) to the prediction model to receive a prediction.
No machine learning model is perfect. It is quite common to retrain CNNs on new data, including updated labels for misclassified images. The Cancer Prediction System allows for retraining and automates the process with Kubeflow Pipelines.
6 Deploy the applications #
Cancer Prediction System is built on top of Flask, a lightweight Web application framework.
You can find the code under application inside your project directory, which you created earlier by cloning the project repository.
The solution contains two separate applications: one for the doctor (under doctor_app) and one the radiologist (under lab_tech).
/application
-- doctor_app/
-- app.py
-- Dockerfile
-- classification-model.h5
-- prediction-model.h5
-- requirements.txt
-- static/
-- styles/
-- css/
-- js/
-- template/
-- base.html
-- gallery.html
-- predict.html
-- retrain.html
-- upload.html
-- lab_tech/
-- app.py
-- Dockerfile
-- classification-model.h5
-- adjust.py
-- requirements.txt
-- static/
-- styles/
-- css/
-- js/
-- template/
-- base.html
-- predict.html
-- send.html
-- upload.html6.1 Deploy and access the Lab Technician Interface #
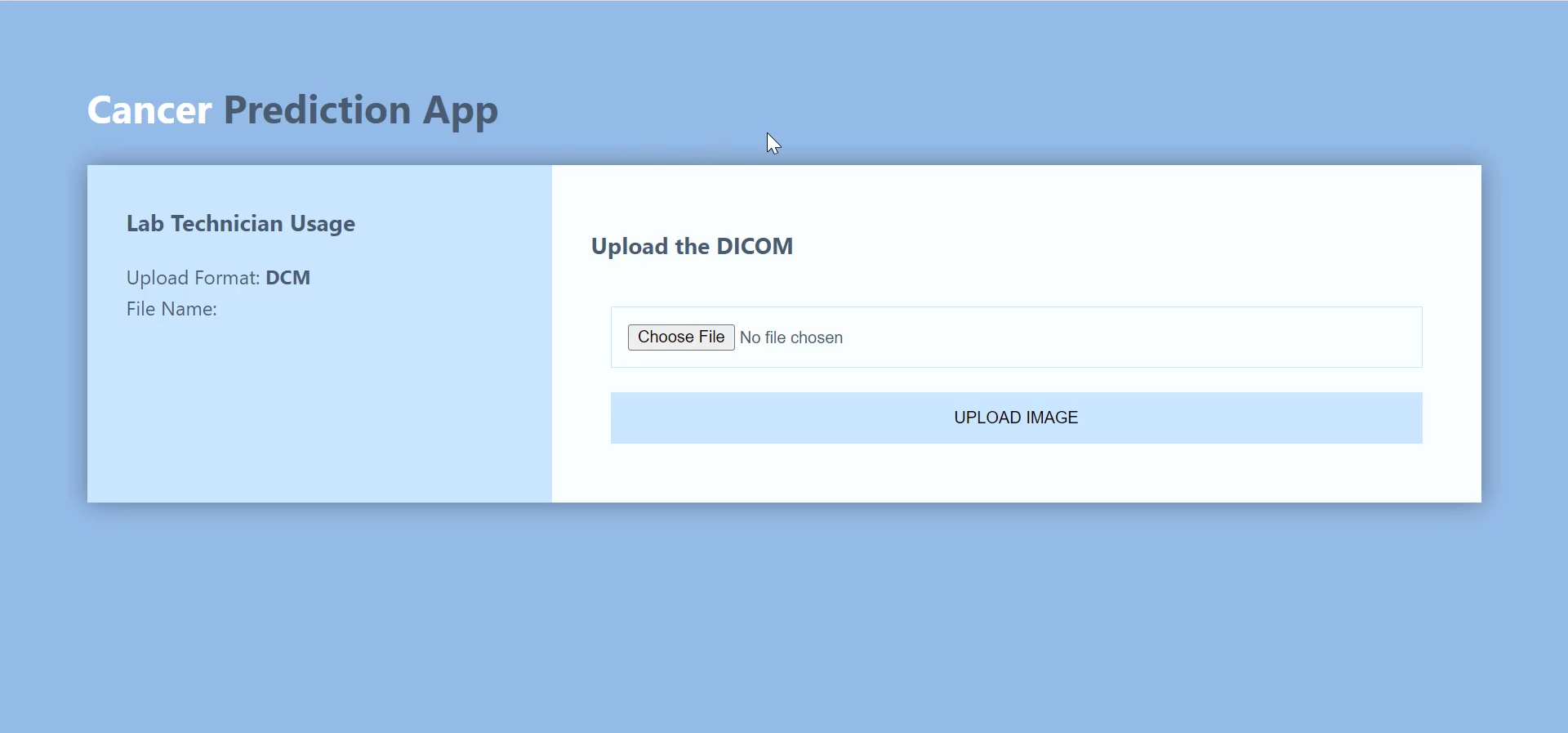
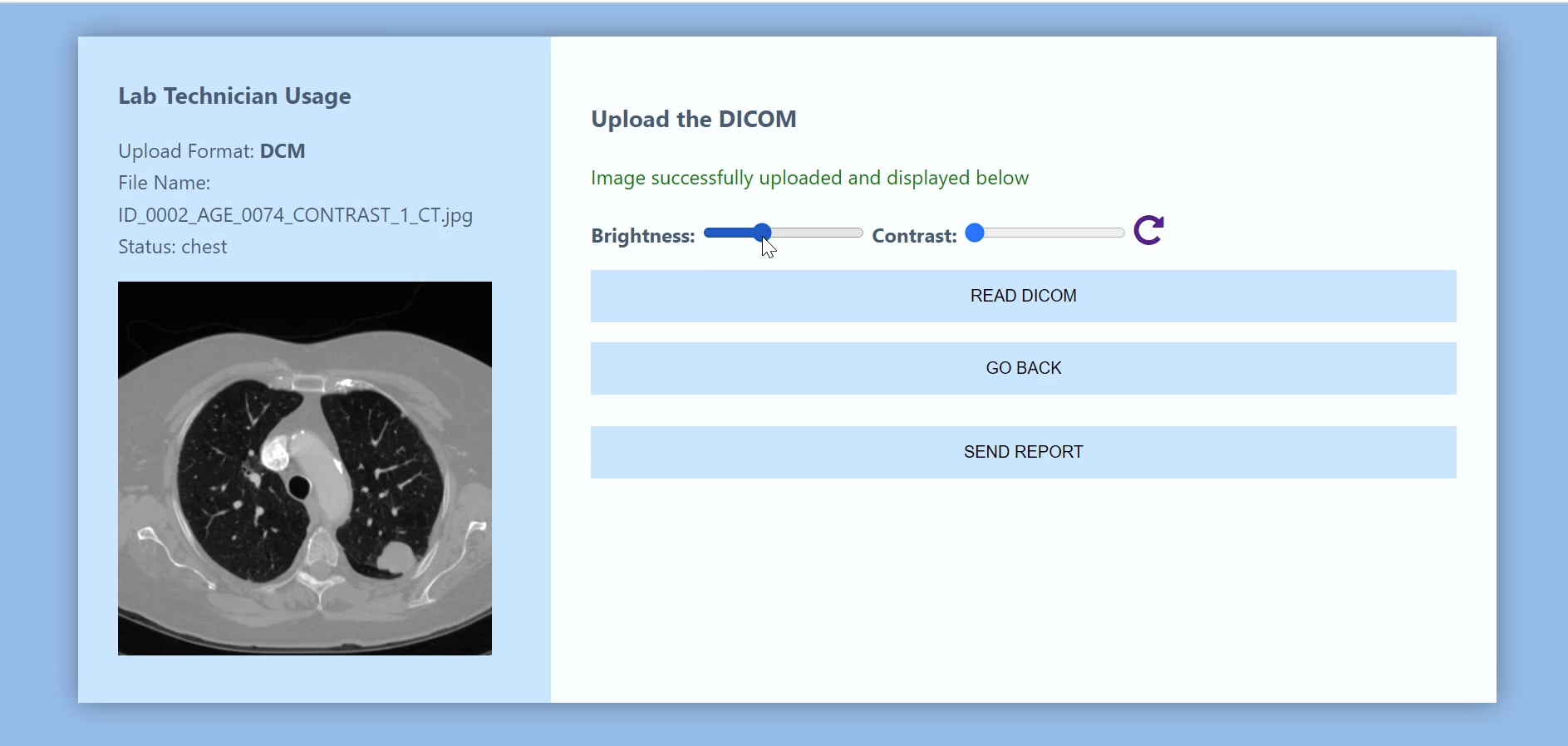
The Lab Technician Interface is responsible for getting DICOM image as input. The person (radiologist, lab technician, physician) can read all the information associated with the DICOM image and alter information, such as contrast, brightness, or angle of rotation.
The Lab Technician Interface has a streamlined installation process using Kubernetes manifests.
If you have not already done so, clone the application code repository.
git clone https://github.com/abhi-bhatra/ct_image_scanning.git
Change to the
ct_image_scanning/kubernetes-manifests/lab-techdirectory.cd ct_image_scanning/kubernetes-manifests/lab-tech
Apply the kustomization file to deploy the Lab Technician Interface
kubectl apply -k .
6.2 Deploy the Doctor’s Dashboard #
The Doctor’s Dashboard lets a doctor access reports sent by a lab technician, along with the Cancer Prediction System’s cancer prediction.
You can install the Doctor’s Dashboard with the manifests included in the code repository that you cloned in the previous section.
Change the directory to
ct_image_scanning/kubernetes-manifests/doctor-app/Apply the kustomization file to deploy the Doctor’s Dashboard Interface
kubectl apply -k .
6.3 Verify installation #
After you perform the above installations, validate that the Cancer Prediction System is installed and running.
List the pods running in the 'cancerns' namespace.
kubectl get all --namespace cancerns
This should produce output like the following:
NAME READY STATUS RESTARTS AGE pod/datasetvm-5db64d7549-cflmf 1/1 Running 0 9s pod/doctor-app-d5b856997-64gnt 1/1 Running 0 10s pod/labtech-app-6c54f58874-jdmcw 1/1 Running 0 12s NAME TYPE CLUSTER-IP EXTERNAL-IP PORT(S) AGE service/doctor-svc NodePort 10.0.66.40 None 5002:30001/TCP 23s service/labtech-svc NodePort 10.0.145.57 None 5001:32009/TCP 24s NAME READY UP-TO-DATE AVAILABLE AGE deployment.apps/datasetvm 1/1 1 0 13s deployment.apps/doctor-app 1/1 1 0 23s deployment.apps/labtech-app 1/1 1 0 23s NAME DESIRED CURRENT READY AGE replicaset.apps/datasetvm-5db64d7549 1 1 1 13s replicaset.apps/doctor-app-d5b856997 1 1 1 10s replicaset.apps/labtech-app-6c54f58874 1 1 1 21s
Verify that you see
doctor-app-andlabtech-app-pods in the 'Running' state and theservice/doctor-svcandservice/labtech-svcare assigned CLUSTER-IP addresses.Connect to
labtech-svcwith your Web browser through the Rancher UI.In the Rancher UI, select the application cluster, then click Services.
Click the displayed port number for the
labtech-svcservice to be redirected to the Lab Technician Interface.
Repeat these steps to connect to the
doctor-svcservice through the Rancher UI.
6.4 Configure external access #
The final step in configuring the Cancer Prediction System is to enable external access.
With SUSE Rancher, you can configure load balancers or ingress controllers to redirect service requests. Load balancers can only handle one IP address per service, while ingress works with one or more ingress controllers to dynamically route service requests.
For real-world use, it is recommended that you configure your cluster with an ingress.
Below is a sample ingress resource for the Cancer Prediction System:
apiVersion: networking.k8s.io/v1
kind: Ingress
metadata:
name: ingress-cancerprediction
spec:
rules:
- host: "cancerpred.example.com"
http:
paths:
- path: /doctor
pathType: Prefix
backend:
service:
name: doctor-svc
port:
number: 443
- path: /labtech
pathType: Prefix
backend:
service:
name: labtech-svc
port:
number: 443You must have properly configured DNS resolution to use a domain name (such as 'cancerpred.example.com') for application access.
7 Operational overview #
The Cancer Prediction System has two primary interfaces, the Lab Technician Interface and the Doctor’s Dashboard.
7.1 Lab Technician Interface #
Open your Web browser to the Lab Technician Interface at 'https://HOSTADDRESS/labtech' (replace HOSTADDRESS with the domain name or IP address you specified in your ingress configuration).
TipYou can still access the Lab Technician Interface through the Rancher UI as you did earlier.
With the Lab Technician Interface, you can upload a new DICOM image.
Adjust brightness and contrast, and then send a report.
7.2 Doctor’s Dashboard #
Open your Web browser to the Doctor’s Dashboard at 'https://HOSTADDRESS/doctor' (replace HOSTADDRESS with the address you defined in your ingress) or through the Rancher UI.
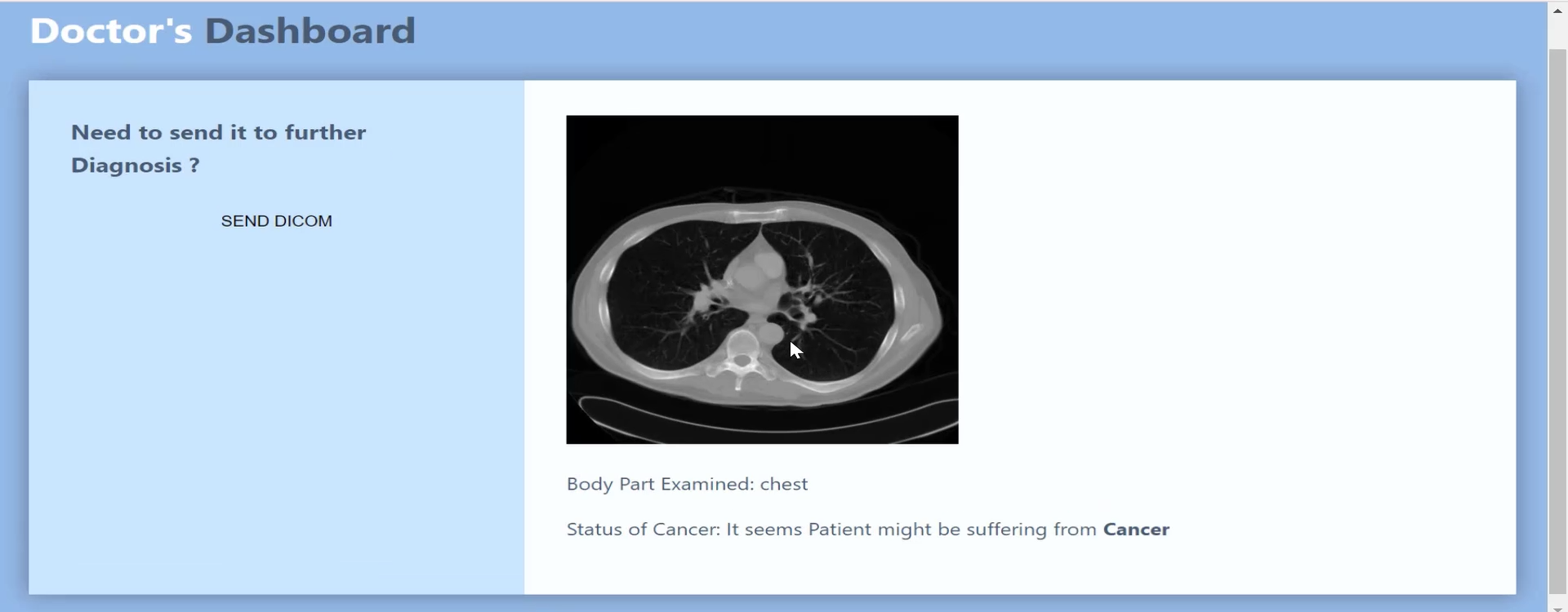
From the Doctor’s Dashboard, the doctor can see and select from available reports.
A report in the Doctor’s Dashboard includes the image, the body part shown, and a cancer prediction.
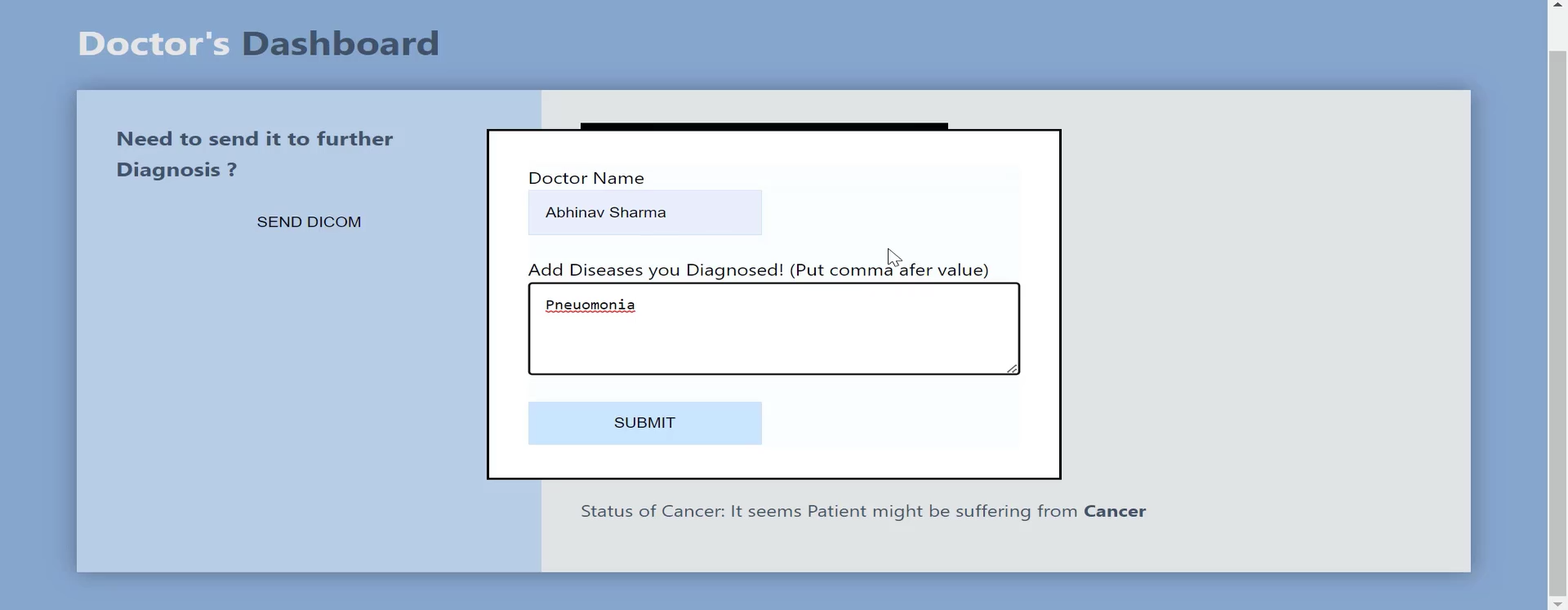
If the doctor is not satisfied with the prediction, the doctor can send the DICOM image back to retrain the model.
8 Summary #
Advanced analytics and machine learning are helping deliver better healthcare outcomes. By leveraging SUSE Rancher and a Kubernetes-native approach, life-saving applications can be deployed everywhere - in the data center, in the cloud, and even at the edge.
Continue your learning journey:
9 Legal notice #
Copyright © 2006–2024 SUSE LLC and contributors. All rights reserved.
Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or (at your option) version 1.3; with the Invariant Section being this copyright notice and license. A copy of the license version 1.2 is included in the section entitled "GNU Free Documentation License".
SUSE, the SUSE logo and YaST are registered trademarks of SUSE LLC in the United States and other countries. For SUSE trademarks, see https://www.suse.com/company/legal/.
Linux is a registered trademark of Linus Torvalds. All other names or trademarks mentioned in this document may be trademarks or registered trademarks of their respective owners.
Documents published as part of the series SUSE Technical Reference Documentation have been contributed voluntarily by SUSE employees and third parties. They are meant to serve as examples of how particular actions can be performed. They have been compiled with utmost attention to detail. However, this does not guarantee complete accuracy. SUSE cannot verify that actions described in these documents do what is claimed or whether actions described have unintended consequences. SUSE LLC, its affiliates, the authors, and the translators may not be held liable for possible errors or the consequences thereof.
10 GNU Free Documentation License #
Copyright © 2000, 2001, 2002 Free Software Foundation, Inc. 51 Franklin St, Fifth Floor, Boston, MA 02110-1301 USA. Everyone is permitted to copy and distribute verbatim copies of this license document, but changing it is not allowed.
0. PREAMBLE#
The purpose of this License is to make a manual, textbook, or other functional and useful document "free" in the sense of freedom: to assure everyone the effective freedom to copy and redistribute it, with or without modifying it, either commercially or noncommercially. Secondarily, this License preserves for the author and publisher a way to get credit for their work, while not being considered responsible for modifications made by others.
This License is a kind of "copyleft", which means that derivative works of the document must themselves be free in the same sense. It complements the GNU General Public License, which is a copyleft license designed for free software.
We have designed this License in order to use it for manuals for free software, because free software needs free documentation: a free program should come with manuals providing the same freedoms that the software does. But this License is not limited to software manuals; it can be used for any textual work, regardless of subject matter or whether it is published as a printed book. We recommend this License principally for works whose purpose is instruction or reference.
1. APPLICABILITY AND DEFINITIONS#
This License applies to any manual or other work, in any medium, that contains a notice placed by the copyright holder saying it can be distributed under the terms of this License. Such a notice grants a world-wide, royalty-free license, unlimited in duration, to use that work under the conditions stated herein. The "Document", below, refers to any such manual or work. Any member of the public is a licensee, and is addressed as "you". You accept the license if you copy, modify or distribute the work in a way requiring permission under copyright law.
A "Modified Version" of the Document means any work containing the Document or a portion of it, either copied verbatim, or with modifications and/or translated into another language.
A "Secondary Section" is a named appendix or a front-matter section of the Document that deals exclusively with the relationship of the publishers or authors of the Document to the Document’s overall subject (or to related matters) and contains nothing that could fall directly within that overall subject. (Thus, if the Document is in part a textbook of mathematics, a Secondary Section may not explain any mathematics.) The relationship could be a matter of historical connection with the subject or with related matters, or of legal, commercial, philosophical, ethical or political position regarding them.
The "Invariant Sections" are certain Secondary Sections whose titles are designated, as being those of Invariant Sections, in the notice that says that the Document is released under this License. If a section does not fit the above definition of Secondary then it is not allowed to be designated as Invariant. The Document may contain zero Invariant Sections. If the Document does not identify any Invariant Sections then there are none.
The "Cover Texts" are certain short passages of text that are listed, as Front-Cover Texts or Back-Cover Texts, in the notice that says that the Document is released under this License. A Front-Cover Text may be at most 5 words, and a Back-Cover Text may be at most 25 words.
A "Transparent" copy of the Document means a machine-readable copy, represented in a format whose specification is available to the general public, that is suitable for revising the document straightforwardly with generic text editors or (for images composed of pixels) generic paint programs or (for drawings) some widely available drawing editor, and that is suitable for input to text formatters or for automatic translation to a variety of formats suitable for input to text formatters. A copy made in an otherwise Transparent file format whose markup, or absence of markup, has been arranged to thwart or discourage subsequent modification by readers is not Transparent. An image format is not Transparent if used for any substantial amount of text. A copy that is not "Transparent" is called "Opaque".
Examples of suitable formats for Transparent copies include plain ASCII without markup, Texinfo input format, LaTeX input format, SGML or XML using a publicly available DTD, and standard-conforming simple HTML, PostScript or PDF designed for human modification. Examples of transparent image formats include PNG, XCF and JPG. Opaque formats include proprietary formats that can be read and edited only by proprietary word processors, SGML or XML for which the DTD and/or processing tools are not generally available, and the machine-generated HTML, PostScript or PDF produced by some word processors for output purposes only.
The "Title Page" means, for a printed book, the title page itself, plus such following pages as are needed to hold, legibly, the material this License requires to appear in the title page. For works in formats which do not have any title page as such, "Title Page" means the text near the most prominent appearance of the work’s title, preceding the beginning of the body of the text.
A section "Entitled XYZ" means a named subunit of the Document whose title either is precisely XYZ or contains XYZ in parentheses following text that translates XYZ in another language. (Here XYZ stands for a specific section name mentioned below, such as "Acknowledgements", "Dedications", "Endorsements", or "History".) To "Preserve the Title" of such a section when you modify the Document means that it remains a section "Entitled XYZ" according to this definition.
The Document may include Warranty Disclaimers next to the notice which states that this License applies to the Document. These Warranty Disclaimers are considered to be included by reference in this License, but only as regards disclaiming warranties: any other implication that these Warranty Disclaimers may have is void and has no effect on the meaning of this License.
2. VERBATIM COPYING#
You may copy and distribute the Document in any medium, either commercially or noncommercially, provided that this License, the copyright notices, and the license notice saying this License applies to the Document are reproduced in all copies, and that you add no other conditions whatsoever to those of this License. You may not use technical measures to obstruct or control the reading or further copying of the copies you make or distribute. However, you may accept compensation in exchange for copies. If you distribute a large enough number of copies you must also follow the conditions in section 3.
You may also lend copies, under the same conditions stated above, and you may publicly display copies.
3. COPYING IN QUANTITY#
If you publish printed copies (or copies in media that commonly have printed covers) of the Document, numbering more than 100, and the Document’s license notice requires Cover Texts, you must enclose the copies in covers that carry, clearly and legibly, all these Cover Texts: Front-Cover Texts on the front cover, and Back-Cover Texts on the back cover. Both covers must also clearly and legibly identify you as the publisher of these copies. The front cover must present the full title with all words of the title equally prominent and visible. You may add other material on the covers in addition. Copying with changes limited to the covers, as long as they preserve the title of the Document and satisfy these conditions, can be treated as verbatim copying in other respects.
If the required texts for either cover are too voluminous to fit legibly, you should put the first ones listed (as many as fit reasonably) on the actual cover, and continue the rest onto adjacent pages.
If you publish or distribute Opaque copies of the Document numbering more than 100, you must either include a machine-readable Transparent copy along with each Opaque copy, or state in or with each Opaque copy a computer-network location from which the general network-using public has access to download using public-standard network protocols a complete Transparent copy of the Document, free of added material. If you use the latter option, you must take reasonably prudent steps, when you begin distribution of Opaque copies in quantity, to ensure that this Transparent copy will remain thus accessible at the stated location until at least one year after the last time you distribute an Opaque copy (directly or through your agents or retailers) of that edition to the public.
It is requested, but not required, that you contact the authors of the Document well before redistributing any large number of copies, to give them a chance to provide you with an updated version of the Document.
4. MODIFICATIONS#
You may copy and distribute a Modified Version of the Document under the conditions of sections 2 and 3 above, provided that you release the Modified Version under precisely this License, with the Modified Version filling the role of the Document, thus licensing distribution and modification of the Modified Version to whoever possesses a copy of it. In addition, you must do these things in the Modified Version:
Use in the Title Page (and on the covers, if any) a title distinct from that of the Document, and from those of previous versions (which should, if there were any, be listed in the History section of the Document). You may use the same title as a previous version if the original publisher of that version gives permission.
List on the Title Page, as authors, one or more persons or entities responsible for authorship of the modifications in the Modified Version, together with at least five of the principal authors of the Document (all of its principal authors, if it has fewer than five), unless they release you from this requirement.
State on the Title page the name of the publisher of the Modified Version, as the publisher.
Preserve all the copyright notices of the Document.
Add an appropriate copyright notice for your modifications adjacent to the other copyright notices.
Include, immediately after the copyright notices, a license notice giving the public permission to use the Modified Version under the terms of this License, in the form shown in the Addendum below.
Preserve in that license notice the full lists of Invariant Sections and required Cover Texts given in the Document’s license notice.
Include an unaltered copy of this License.
Preserve the section Entitled "History", Preserve its Title, and add to it an item stating at least the title, year, new authors, and publisher of the Modified Version as given on the Title Page. If there is no section Entitled "History" in the Document, create one stating the title, year, authors, and publisher of the Document as given on its Title Page, then add an item describing the Modified Version as stated in the previous sentence.
Preserve the network location, if any, given in the Document for public access to a Transparent copy of the Document, and likewise the network locations given in the Document for previous versions it was based on. These may be placed in the "History" section. You may omit a network location for a work that was published at least four years before the Document itself, or if the original publisher of the version it refers to gives permission.
For any section Entitled "Acknowledgements" or "Dedications", Preserve the Title of the section, and preserve in the section all the substance and tone of each of the contributor acknowledgements and/or dedications given therein.
Preserve all the Invariant Sections of the Document, unaltered in their text and in their titles. Section numbers or the equivalent are not considered part of the section titles.
Delete any section Entitled "Endorsements". Such a section may not be included in the Modified Version.
Do not retitle any existing section to be Entitled "Endorsements" or to conflict in title with any Invariant Section.
Preserve any Warranty Disclaimers.
If the Modified Version includes new front-matter sections or appendices that qualify as Secondary Sections and contain no material copied from the Document, you may at your option designate some or all of these sections as invariant. To do this, add their titles to the list of Invariant Sections in the Modified Version’s license notice. These titles must be distinct from any other section titles.
You may add a section Entitled "Endorsements", provided it contains nothing but endorsements of your Modified Version by various parties—for example, statements of peer review or that the text has been approved by an organization as the authoritative definition of a standard.
You may add a passage of up to five words as a Front-Cover Text, and a passage of up to 25 words as a Back-Cover Text, to the end of the list of Cover Texts in the Modified Version. Only one passage of Front-Cover Text and one of Back-Cover Text may be added by (or through arrangements made by) any one entity. If the Document already includes a cover text for the same cover, previously added by you or by arrangement made by the same entity you are acting on behalf of, you may not add another; but you may replace the old one, on explicit permission from the previous publisher that added the old one.
The author(s) and publisher(s) of the Document do not by this License give permission to use their names for publicity for or to assert or imply endorsement of any Modified Version.
5. COMBINING DOCUMENTS#
You may combine the Document with other documents released under this License, under the terms defined in section 4 above for modified versions, provided that you include in the combination all of the Invariant Sections of all of the original documents, unmodified, and list them all as Invariant Sections of your combined work in its license notice, and that you preserve all their Warranty Disclaimers.
The combined work need only contain one copy of this License, and multiple identical Invariant Sections may be replaced with a single copy. If there are multiple Invariant Sections with the same name but different contents, make the title of each such section unique by adding at the end of it, in parentheses, the name of the original author or publisher of that section if known, or else a unique number. Make the same adjustment to the section titles in the list of Invariant Sections in the license notice of the combined work.
In the combination, you must combine any sections Entitled "History" in the various original documents, forming one section Entitled "History"; likewise combine any sections Entitled "Acknowledgements", and any sections Entitled "Dedications". You must delete all sections Entitled "Endorsements".
6. COLLECTIONS OF DOCUMENTS#
You may make a collection consisting of the Document and other documents released under this License, and replace the individual copies of this License in the various documents with a single copy that is included in the collection, provided that you follow the rules of this License for verbatim copying of each of the documents in all other respects.
You may extract a single document from such a collection, and distribute it individually under this License, provided you insert a copy of this License into the extracted document, and follow this License in all other respects regarding verbatim copying of that document.
7. AGGREGATION WITH INDEPENDENT WORKS#
A compilation of the Document or its derivatives with other separate and independent documents or works, in or on a volume of a storage or distribution medium, is called an "aggregate" if the copyright resulting from the compilation is not used to limit the legal rights of the compilation’s users beyond what the individual works permit. When the Document is included in an aggregate, this License does not apply to the other works in the aggregate which are not themselves derivative works of the Document.
If the Cover Text requirement of section 3 is applicable to these copies of the Document, then if the Document is less than one half of the entire aggregate, the Document’s Cover Texts may be placed on covers that bracket the Document within the aggregate, or the electronic equivalent of covers if the Document is in electronic form. Otherwise they must appear on printed covers that bracket the whole aggregate.
8. TRANSLATION#
Translation is considered a kind of modification, so you may distribute translations of the Document under the terms of section 4. Replacing Invariant Sections with translations requires special permission from their copyright holders, but you may include translations of some or all Invariant Sections in addition to the original versions of these Invariant Sections. You may include a translation of this License, and all the license notices in the Document, and any Warranty Disclaimers, provided that you also include the original English version of this License and the original versions of those notices and disclaimers. In case of a disagreement between the translation and the original version of this License or a notice or disclaimer, the original version will prevail.
If a section in the Document is Entitled "Acknowledgements", "Dedications", or "History", the requirement (section 4) to Preserve its Title (section 1) will typically require changing the actual title.
9. TERMINATION#
You may not copy, modify, sublicense, or distribute the Document except as expressly provided for under this License. Any other attempt to copy, modify, sublicense or distribute the Document is void, and will automatically terminate your rights under this License. However, parties who have received copies, or rights, from you under this License will not have their licenses terminated so long as such parties remain in full compliance.
10. FUTURE REVISIONS OF THIS LICENSE#
The Free Software Foundation may publish new, revised versions of the GNU Free Documentation License from time to time. Such new versions will be similar in spirit to the present version, but may differ in detail to address new problems or concerns. See http://www.gnu.org/copyleft/.
Each version of the License is given a distinguishing version number. If the Document specifies that a particular numbered version of this License "or any later version" applies to it, you have the option of following the terms and conditions either of that specified version or of any later version that has been published (not as a draft) by the Free Software Foundation. If the Document does not specify a version number of this License, you may choose any version ever published (not as a draft) by the Free Software Foundation.
ADDENDUM: How to use this License for your documents#
Copyright (c) YEAR YOUR NAME. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any later version published by the Free Software Foundation; with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts. A copy of the license is included in the section entitled “GNU Free Documentation License”.
If you have Invariant Sections, Front-Cover Texts and Back-Cover Texts, replace the “ with…Texts.” line with this:
with the Invariant Sections being LIST THEIR TITLES, with the Front-Cover Texts being LIST, and with the Back-Cover Texts being LIST.
If you have Invariant Sections without Cover Texts, or some other combination of the three, merge those two alternatives to suit the situation.
If your document contains nontrivial examples of program code, we recommend releasing these examples in parallel under your choice of free software license, such as the GNU General Public License, to permit their use in free software.